

 |
 |
Comp Chem Research Developments | |
| Archive of Comp Chem Research News | |
July 26, 2000
|
|
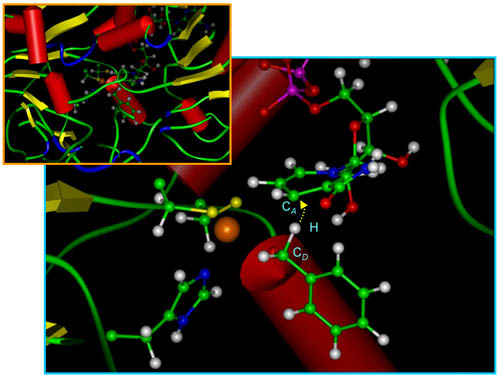
Many enzyme reactions involve proton or hydride transfer and can be expected to proceed by quantum mechanical tunneling. Although great progress has been made in incorporating quantum effects into gas-phase reactions, most simulations of processes involving proteins have involved classical mechanics, and therefore they have been unable to properly model proton and hydride transfer processes. This has been particularly frustrating because kinetic isotope effects are very sensitive to tunneling, and kinetic isotope effects are often the best means for learning about transition state structure. This situation has now changed dramatically, as reported in a paper accepted for publication recently in Journal of the American Chemical Society. The paper is authored by Professors Jiali Gao and Donald G. Truhlar of the Chemistry Department along with three research associates in the department, Drs. Cristobal Alhambra, José Corchado, and Maria Luz Sánchez, and it reports a simulation of the reaction rates and kinetic isotope effects of the hydride transfer for benzyl alcoholate anion to the coenzyme NAD+, catalyzed by the enzyme liver alcohol dehydrogenase. The calculation was made possible by two advances in simulation methods. First is the treatment of the force field, which involves a combination of semiempirical molecular orbital theory, semiempirical valence bond terms, and molecular mechanics. Second is the treatment of atomic motions, which is based on variational transition state theory with quantized vibrations and multidimensional tunneling contributions along optimized tunneling paths. The calculations agree very well with kinetic isotope effects measured by Professor Judith Klinman and coworkers at Berkeley, and they provide an interpretation of the highly nonclassical kinetic isotope effects that they observed in terms of the rehybridization at the donor carbon atom. This can be seen clearly in the figure, where the hybridization of this carbon atom, caught in the process of releasing the tunneling hydride atom, is clearly intermediate between sp2 and sp3. |
|
 |
|
|
| |