

 |
 |
Comp Chem Research Developments | |
| Archive of Comp Chem Research News | |
|
September 29, 2004 | |
| |
|
The research group of Prof. Darrin York of the Department of Chemistry has
recently made several significant advances in the development multi-scale quantum
models to study the molecular mechanisms of RNA catalysis.
These models involve the integration of a hierarchy of theoretical levels
that work together synchronously to provide detailed insight into complex
biological processes that simultaneously span a broad range of spatial and
temporal domains. An understanding of the mechanisms of phosphoryl
transfer reactions involved in RNA catalysis is important for the design
of new medical therapies that target genetic disorders as well as the
development of new biotechnology such as RNA chips. The York
Group has has constructed a large-scale density-functional quantum
database of model phosphoryl transfer reactions (in the gas phase and with
continuum solvent corrections). The QCRNA database
has recently gone online and contains over 1,500 molecular structures and
200 chemical mechanisms, and represents the world's largest database of biochemical
phosphoryl transfer reactions. Recent studies resulting from the
data contained in QCRNA have been
published [J. Am. Chem.
Soc., 126, 1654 (2004);
Chem. Phys. Chem., 5, 1045 (2004); J. Biol. Inorg.
Chem. DOI:10.1007/ s00775-
004-0583-7]. | |
(Click on the image to view
a 2.0MB MPEG movie)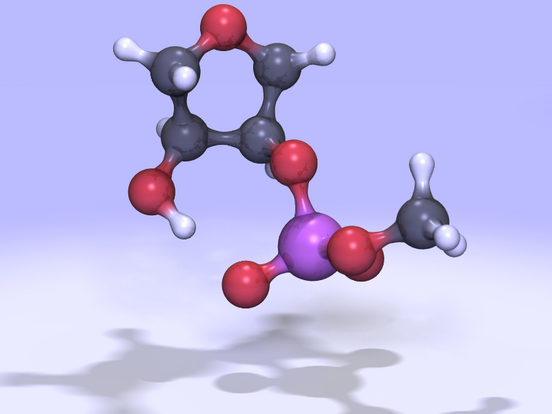
| |
| Figure 1: A
5'-ribose,3'-phosphodiester model for RNA transesterification and
hydrolysis. Movies of the reaction can be viewed as a 2.0MB MPEG file or 10MB animated GIF file. | |
|
The QCRNA
database has been used to design new fast semiempirical quantum models
that can be used in simulations of model RNA catalysis reactions in
complex chemical environments. Combined quantum mechanical/molecular
mechanical (QM/MM) simulations have recently explored the nature of
transphosphorylation thio effects (the change in reaction rate that occurs
upon substitution of key phosphoryl oxygen positions with sulfur) to aid
in mechanistic interpretation of experimental results [J. Am. Chem. Soc., 126, 7504 (2004)]. The York
Group has recently extended the QM/MM methodology to include a new smooth COSMO solvation method for biological reactions,
a variational electrostatic projection method for
efficient modeling of the solvated macromolecular environment in activated
dynamics simulations, and a new efficient linear-scaling Ewald technique for long-range
electrostatic interactions in collaboration with Prof. Jiali
Gao. New-generation semiempirical quantum models derived from QCRNA are
forthcoming, however preliminary quantum models have already emerged such
as the AM1/d* method for phosphate
hydrolysis reactions [Theor. Chem.
Acc. 109, 149
(2003)], and the PM3BP method for nucleotide base
pairing [J. Comput.
Chem. 24, 57
(2003)] done in collaboration with the group of Prof. Christopher
Cramer. These models can be used with linear-scaling electronic
structure methods, also developed by the York Group, to examine new
quantum descriptors for entire solvated biological macromolecules up to
tens of thousands of atoms [Proteins, 56, 724-737 (2004)]. This
strategy has recently been applied to study the regioselectivity and RNA
binding affinity of the HIV-1 nucleocapsid protein [J. Mol. Biol. 330,
993 (2003)] in collaboration with the group of Prof.
Karin Musier-Forsyth. | |
|
| |