

 |
 |
Comp Chem Research Developments | |
| Archive of Comp Chem Research News | |
June 12, 2002
|
|
|
|
The study of catalytic RNA systems is fundamental to biology. Indeed, the recent discovery that the catalytic core of the cellular ribosome is predominantly RNA underscores the importance of RNA catalysis and lends credence to the "RNA World" hypothesis of the origins of life. The manipulation of chemistry of catalytic RNAs (or "ribozymes") holds considerable promise for pharmaceutical and biotech industries. Despite the wealth of experimental data available for catalytic RNA systems, the details of the reaction mechanisms remain elusive. If known, these details could help lead to the design of new gene therapies, RNA chips that might be used as diagnostic tools in medicine, or biomolecular switches used in nanodevices. From a computational perspective, RNA presents many challenges related to it's high negative charge, inherent flexibility, interaction with metal ions and d-orbital phosphate chemistry. Recently, a new quantum method has been designed by chemistry professor Darrin York and Minnesota Supercomputing Institute Research Scholar Xabier Lopez. The method, which will appear in an upcoming special issue of Theoretical Chemistry Accounts, allows the calculation of phosphate hydrolysis reactions (Fig. 1) in ribozymes with accuracy comparable to that of high-level density-functional calculations for a thousand-fold reduction in computational cost. The methods can be used with new "linear-scaling" quantum methods for macromolecular characterization developed by professor York and graduate student Jana Khandogin (to appear in an upcoming issue of the Journal of Physical Chemistry) that allows RNA systems of several thousands of atoms to be studies in solution (Fig. 2). |
|
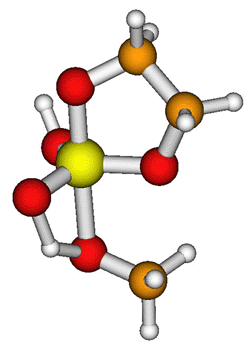 Fig. 1: Transition state in a model phosphate hydrolysis reaction.
|
|
|
| |