

 |
 |
Comp Chem Research Developments | |
| Archive of Comp Chem Research News | |
January 10, 2001
|
|
|
|
|
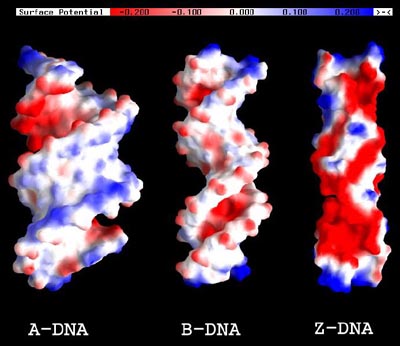
Professor Darrin York with graduate student Jana Khandogin and Minnesota Supercomputing Institute research fellow Anguang Hu have recently developed new methods for the calculation of quantum electronic structure of large biomolecules in solution. These calculations are of tremendous importance for the fields of computational biochemistry. Conventional quantum mechanical methods can be applied only to fairly small systems since the computational cost increases extremely steeply with the system size (for a large system, doubling the system size increases the cost by a factor of 8-16 or even more, depending on the method). The York group has developed new linear-scaling quantum methods that circumvent this "scaling bottleneck" and allow very large systems to be studied with computational quantum models. In a recent special issue of the Journal of Computational Chemistry on "Quantum Chemical Methods for Large Molecules", the York group reports the application of these methods to DNA and RNA systems in solution. The figure below illustrates how different forms of DNA are polarized by solvent - a quantum mechanical effect that cannot be captured by conventional molecular mechanical "force field" models that neglect polarization. The researchers can derive from their calculations new quantum indices for macromolecular characterization such as the polarization density and local chemical softness functions that may prove extremely useful in applications to drug design problems. This works lays the groundwork for ongoing studies in the York group on the reactivity of catalytic RNAs. |
|
 |
|
|
The electrostatic potential map of the solvent-induced polarization response density (the difference between the DNA electron density that is polarized by solvent and the unpolarized density of an isolated DNA molecule) of A, B and Z-form DNA. |
|
|
| |