

 |
 |
Comp Chem Research Developments | |
| Archive of Comp Chem Research News | |
February 7, 2001
|
|
|
|
|
Graduate student Alessandro Mascioni and Prof. Gianluigi Veglia and, in collaboration with Prof. David Thomas and Dr. Christine Karim at the Dept. of Biochemistry, Molecular Biology and Biophysics, have recently determined the structure of the human protein sarcolipin (SLN), that is involved in the regulation of the sarcoplasmic reticulum calcium pump (SERCA). Muscle contraction is initiated by the release of calcium from the sarcoplasmic reticulum, which pumps Ca2+ actively into its lumen by means of the SERCA, also known as Ca-ATPase. Sarcolipin is a 31 amino acid protein, which interacts with the Ca-ATPase (SERCA1a) present in human fast-twitch skeletal muscle and regulates its activity. SLN (Figure 1) was first synthesized by Dr. Christine Karim in Prof. Barany` s laboratory using solid-phase peptide synthesis, and the structure then obtained in Dr. Veglia’s laboratory from proton 2D NOESY experiments performed on the 800MHz Varian Spectrometer available at the NMR facility in the BSBE. The figure below represents the high-resolution structure of the protein obtained by solution NMR in micelle solution. The structure of SLN will be successively used in conjunction with additional NMR experiments to investigate its interaction with Ca-ATPase, expanding our present knowledge about the regulation of the calcium pump in human skeletal muscles. Professor Veglia’s research focus on the structure determination of membrane proteins by solution and solid-state NMR spectroscopy. Other projects currently active in his laboratory include the structure determination of phospholamban, (another well known Ca-ATPase regulator), and the structure of stannin, involved in the toxicity of organotin compounds and apoptosis. |
|
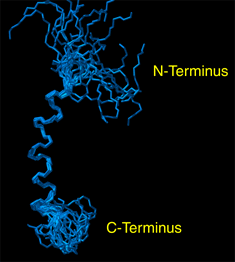 |
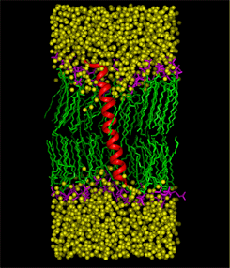 Figure 2 |
Graduate student Ramkumar Rajamani and Professor Jiali Gao have recently developed a structural and dynamic model of sarcolipin, a 31-amino acid transmembrane protein that affects muscle contraction by regulating the Ca+2 affinity of the skeletal muscle sarcoplasmic reticulum Ca+2-ATPase. To understand the mechanism of sarcolipin- Ca+2-ATPase interaction, it is essential to know the three-dimensional structure of the protein and its dynamics in the membrane environment. Concomitant with the theoretical studies, the structure of sarcolipin is also being investigated by Professor Gianluigi Veglia of the Chemistry Department, using NMR spectroscopy. However, it was decided not to compare results between the two groups during the course of the study in order to provide an independent test of the computational methods. A major question on the structure of sarcolipin is the tilt angle between the helical axis of sarcolipin with respect to the normal direction (z) of the membrane. Thus, three separate molecular dynamics simulations of sarcolipin embedded in the DMPC lipid membrane, corresponding to tilt angles of 0o, 10o, and 20o, have been carried out for about 3 ns, using NPT ensemble at 51 oC and 1 atm along with periodic boundary conditions and the particle-mesh Ewald technique for long-range electrostatic interactions. The simulations converged to a tilt angle of about 7-13o, which is in good agreement with the NMR observation of 10o for phospholamban, which is a homologues protein, acting as a major regulator of cardiac contractility. The structure obtained from molecular dynamics simulations is found in accord with the NMR structure determined by Veglia and Mascioni, with a RMSD difference of 1.2 angstroms in the transmembrane region. Another major issue is the dynamic properties of the sarcolipin in the membrane, which is critical in the understanding of sarcolipin-Ca+2-ATPase binding. The lateral diffusion coefficient has been estimated from the molecular dynamics trajectory, and a value of 1.7 x 10-8 cm2/sec is obtained, which may be compared with the diffusion coefficient of 10-7 to 10-8 cm2/sec for phospholipids. The similarity of the two diffusion coefficients is attributed to the formation of hydrogen bonds between the side chains of Arg6 and Arg27 and the lipid head groups. The molecular dynamics simulations were carried out using 16 processors of IBM SP machine at the Minnesota Supercomputing Institute. Figure 2 shows the last structure from a 3-ns molecular dynamics simulation of sarcolipin in the full protein-membrane system. The system consists of the 31-amino acid protein, sarcolipin, 44 DMPC lipid molecules, and 1849 water molecules, totaling 11,289 atoms. Sarcolipin is shown by its secondary structure in red, the lipid tails are given in green and head groups in pink, and water molecules (without hydrogens) are represented in yellow. |
|
|
| |